Help for FungalRV adhesin Predictor
Help for Immunoinformatics Database Search Query Making Options | The informations present in FungalRV Database can be retrieved through the "Immunoinformatics Data" tab which takes user to the Immunoinformatics data query page. The information to be retrieved through this page can be divided into two layers. First Layer having general information of motif, topology, location, homology and antigenic regions and Second Layer having epitope and allergen informations for the predicted adhesins and adhesin-like proteins. The information present in FungalRV database van also be accessed through the "Go to Advanced Search" link.Through this page users can filter their results on the basis of Protein length, number of transmembrane spanning regions, localization and reliability class, presence or absence of betawraps, paralogs, hits to Conserved Domain Database and Human Reference proteins. (I) Query Making Through "Immunoinformatics Data" query page(i) Select a pathogen from the pull down menu list.  (ii) Provide an Orf ID corresponding to the selected pathogen. For example if user selects Aspergillus fumigatus, then the Orf ID provided by the user must correspond to Aspergillus fumigatus. Multiple comma separated ORF IDs can also be provided eg- Afu2g05150,Afu2g10130. This field is optional for first layer data retrieval but must be provided to fetch second layer data. Second layer data can be retrieved corresponding to a single Orf ID against the pathogen selected. Click this link to get the list of ORF Ids corresponding to available "307 predicted adhesins and adhesin-like proteins"  (iii) The user can also retrieve the First Layer information corresponding to a keyword. This field is optional.
|  (iv) Select checkbox corresponding to the data user wants to retrieve in Data Available frame.
| 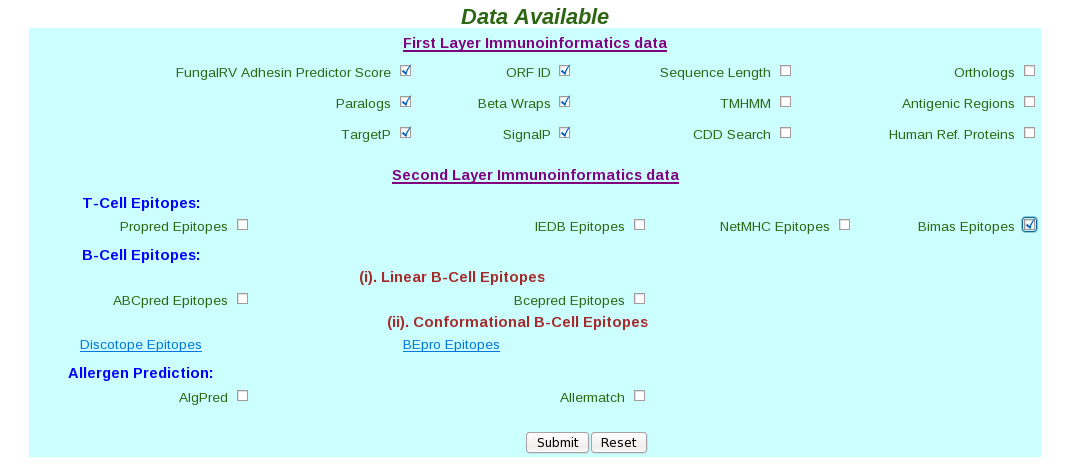 (v) In the Second Layer the Conformational B-Cell Epitope data is available as flat files, these are hyper linked. The flat files can be saved by the file "save as" option by the users for future use.
| (vi) Finally click the submit button. This takes users to the result page.  (vii) In the result page the entire quiried data can be saved by the user on clicking the hyperlink in blue "Export data for First Layer" which provides facility to save the First Layer Data in text file and the successive epitope and allergen hyperlinks "Export Bimas Epitope Data" etc which provides facility to save the Second Layer Data from individual servers in text files. 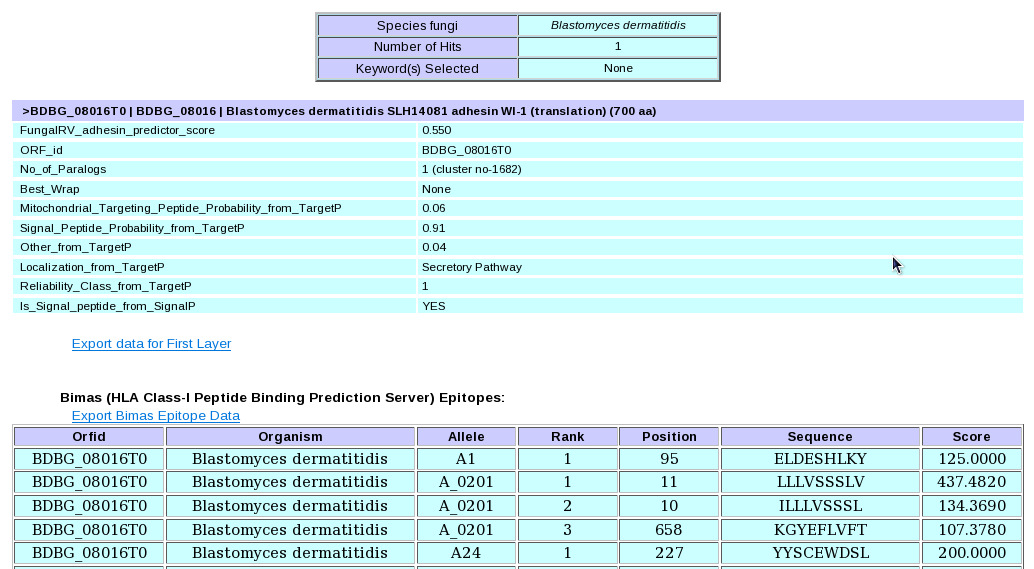 (II) Query Making through FungalRV "Advanced Search" page | (i) Select a pathogen from the pull down menu list. |  (ii) Select checkbox corresponding to the data user wants to retrieve in "Show information for:" frame | 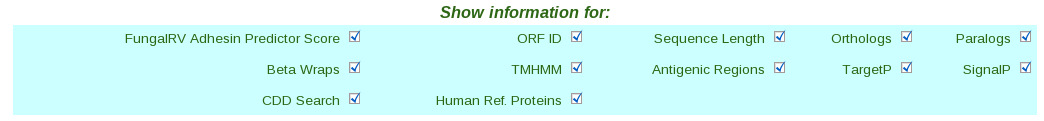 (iii) Set the filters corresponding to the data selected through the drop down boxes. | 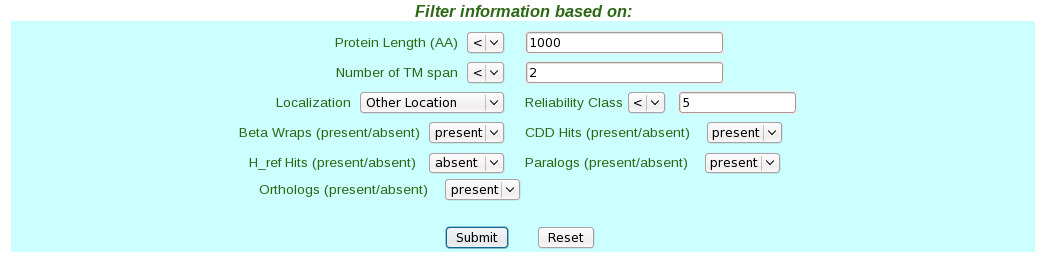 (iv) Finally click the submit button which takes users to the result page. | 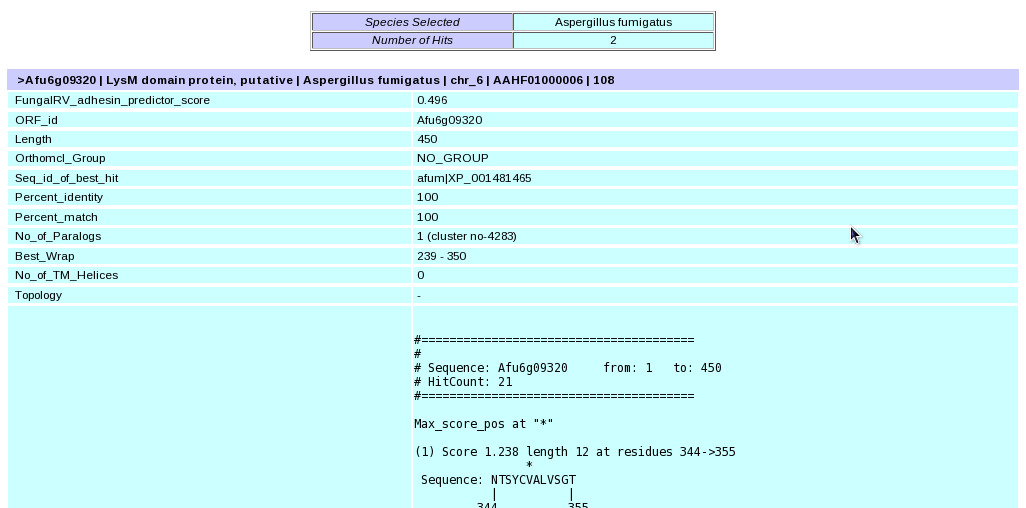 (v) In the result page the entire queried data can be saved by the user on clicking the hyperlink in blue "Export" which provides facility to save the entire queried First Layer Data into a text file. | Viewing Known Vaccines Candidate Information
| (i) The Fungal Known Vaccine Candidate Data can be viewed by clicking the "Known Vaccines" tab. |
FungalRVAdhesin Prediction and Immunoinformatics portal for human fungal pathogens |  |